-
Type:
Story
-
Status: Won't Fix (View Workflow)
-
Priority:
Normal
-
Resolution: Won't Fix
-
Affects Version/s: None
-
Fix Version/s: None
-
Component/s: None
-
Labels:
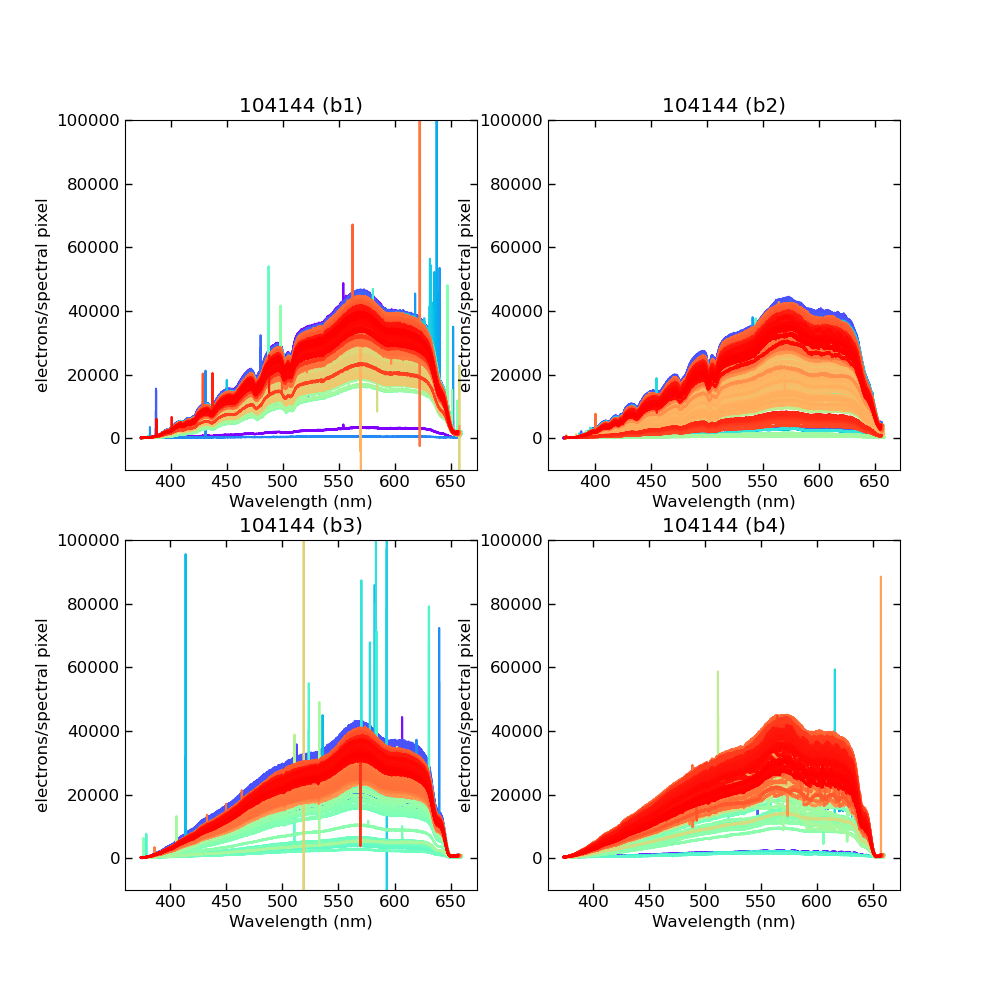
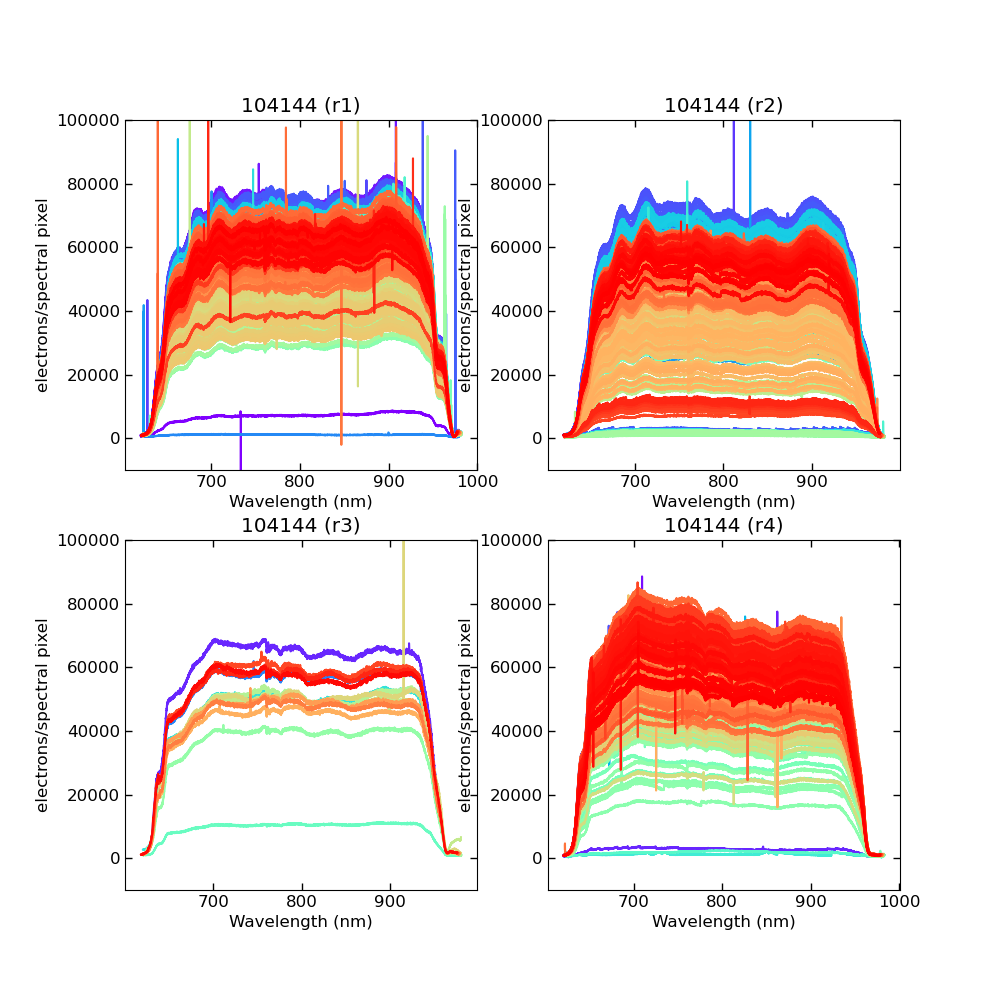
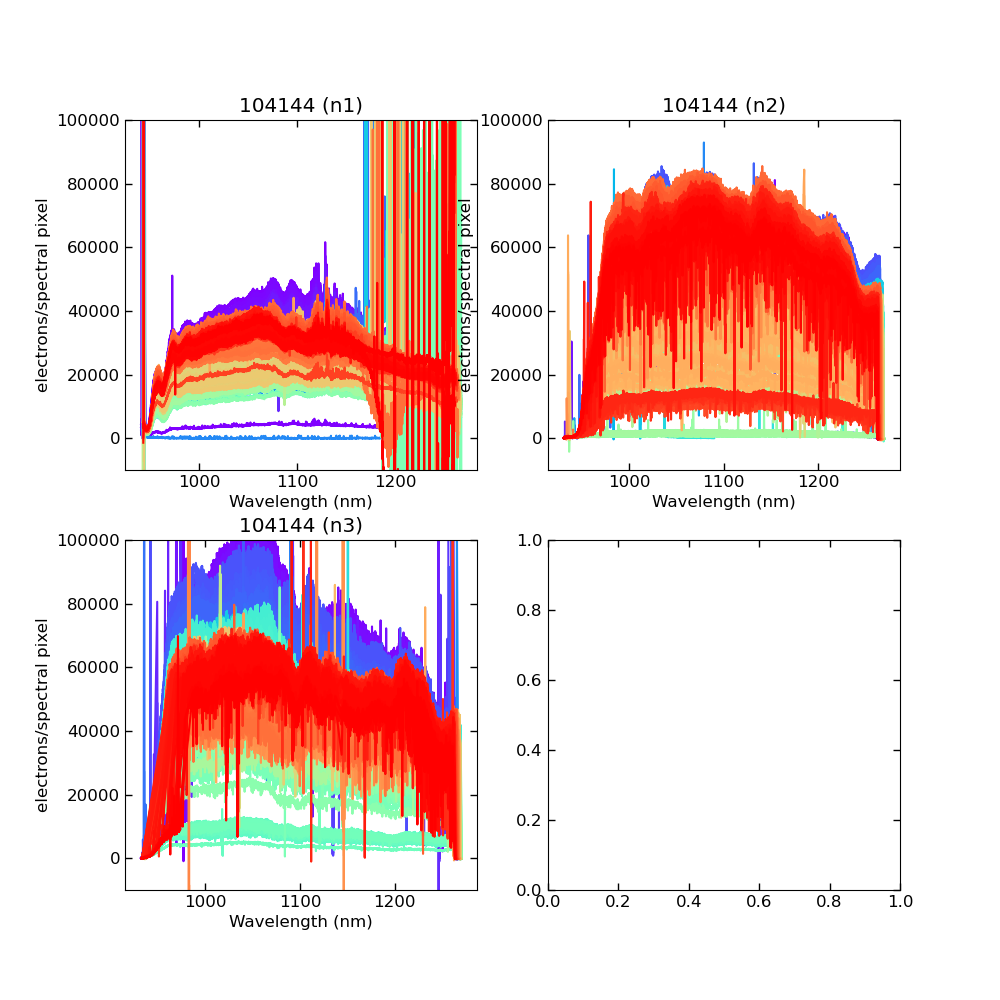
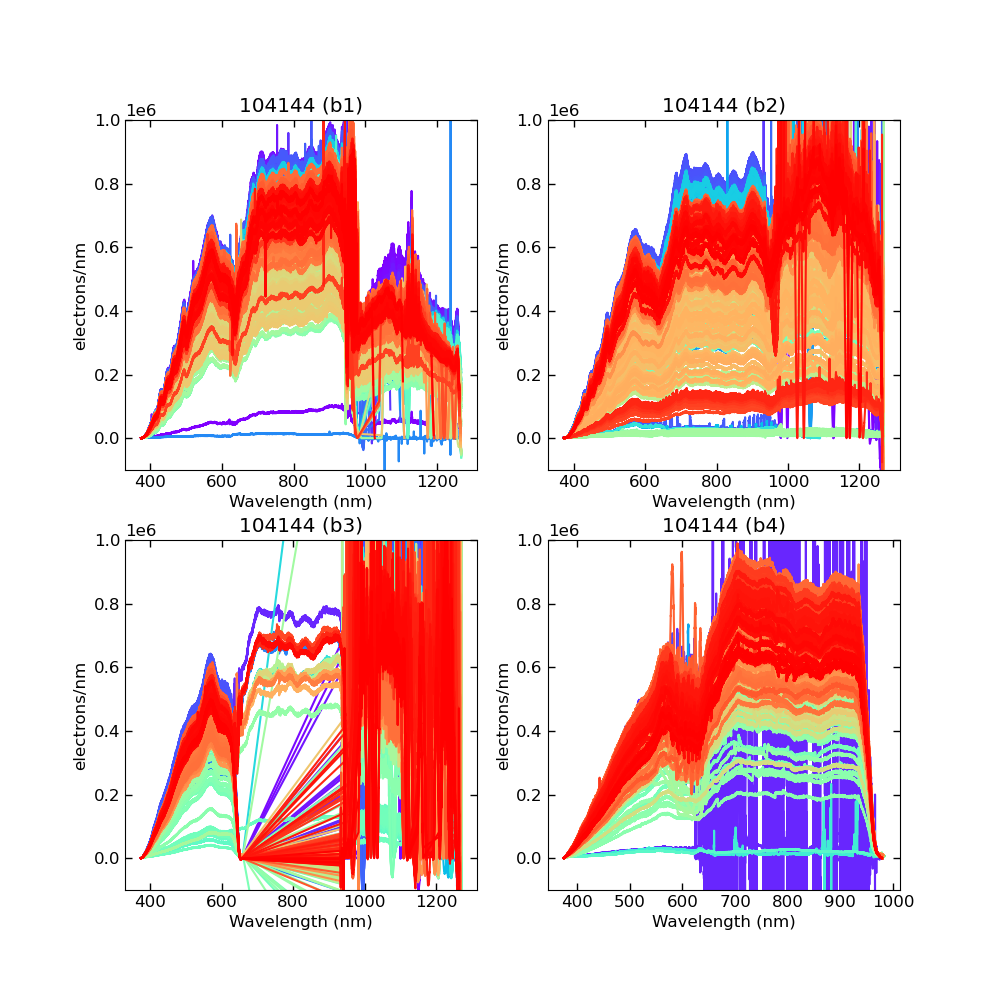
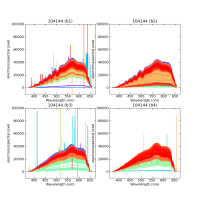
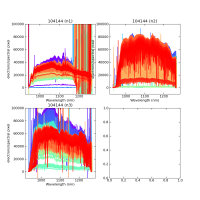
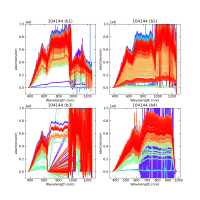
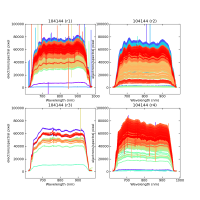
Using CALIBs generated in part of PIPE2D-1346, it looks there are many nan values in many fibers of pfsArm r3. Investigate what is going on.
Here are some examples:
repoDir='/work/drp' pfsDesignDir = '/work/drp/pfsDesign' rerun = os.path.join(repoDir, 'rerun', f'kiyoyabe/calibs/2024jan/20240205') calibRoot = os.path.join('/work/kiyoyabe/drp/calibs/generateCALIBs/2024jan/update_detectorMaps/CALIB-2023-11-v4') butler = dafPersist.Butler(rerun, calibRoot=calibRoot) visit=104144 arm='r' fig = 1; plt.close(fig); fig = plt.figure(fig, figsize=(10,10)) ax1 = fig.add_subplot(221) ax2 = fig.add_subplot(222) ax3 = fig.add_subplot(223) ax4 = fig.add_subplot(224) spectrograph=1 dataId = dict(visit=visit, spectrograph=spectrograph, arm=arm) pfsArm = butler.get('pfsArm', dataId) pfsArm.plot(axes=ax1, ignorePixelMask=256, show=False) ax1.set_ylim(-1e4, 1e5) ax1.set_title(f'{visit} ({arm}{spectrograph})') spectrograph=2 dataId = dict(visit=visit, spectrograph=spectrograph, arm=arm) pfsArm = butler.get('pfsArm', dataId) pfsArm.plot(axes=ax2, ignorePixelMask=256, show=False) ax2.set_ylim(-1e4, 1e5) ax2.set_title(f'{visit} ({arm}{spectrograph})') spectrograph=3 dataId = dict(visit=visit, spectrograph=spectrograph, arm=arm) pfsArm = butler.get('pfsArm', dataId) pfsArm.plot(axes=ax3, ignorePixelMask=256, show=False) ax3.set_ylim(-1e4, 1e5) ax3.set_title(f'{visit} ({arm}{spectrograph})') if True: spectrograph=4 dataId = dict(visit=visit, spectrograph=spectrograph, arm=arm) pfsArm = butler.get('pfsArm', dataId) pfsArm.plot(axes=ax4, ignorePixelMask=256, show=False) ax4.set_ylim(-1e4, 1e5) ax4.set_title(f'{visit} ({arm}{spectrograph})') plt.savefig(f'trace_{arm}.pdf') plt.savefig(f'trace_{arm}.png')
repoDir='/work/drp' pfsDesignDir = '/work/drp/pfsDesign' rerun = os.path.join(repoDir, 'rerun', f'kiyoyabe/calibs/2024jan/20240205') calibRoot = os.path.join('/work/kiyoyabe/drp/calibs/generateCALIBs/2024jan/update_detectorMaps/CALIB-2023-11-v4') butler = dafPersist.Butler(rerun, calibRoot=calibRoot) visit=104144 dataId = dict(visit=visit) pfsMerged = butler.get('pfsMerged', dataId) pfsConfig = butler.get('pfsConfig', dataId) fig = 1; plt.close(fig); fig = plt.figure(fig, figsize=(10,10)) ax1 = fig.add_subplot(221) ax2 = fig.add_subplot(222) ax3 = fig.add_subplot(223) ax4 = fig.add_subplot(224) spectrograph=1 pfsMerged1 =pfsMerged.select(pfsConfig, spectrograph=spectrograph) pfsMerged1.plot(axes=ax1, ignorePixelMask=256, show=False) ax1.set_ylim(-1e5, 1e6) ax1.set_title(f'{visit} (SM{spectrograph})') spectrograph=2 pfsMerged2 =pfsMerged.select(pfsConfig, spectrograph=spectrograph) pfsMerged2.plot(axes=ax2, ignorePixelMask=256, show=False) ax2.set_ylim(-1e5, 1e6) ax2.set_title(f'{visit} (SM{spectrograph})') spectrograph=3 pfsMerged3 =pfsMerged.select(pfsConfig, spectrograph=spectrograph) pfsMerged3.plot(axes=ax3, ignorePixelMask=256, show=False) ax3.set_ylim(-1e5, 1e6) ax3.set_title(f'{visit} (SM{spectrograph})') spectrograph=4 pfsMerged4 =pfsMerged.select(pfsConfig, spectrograph=spectrograph) pfsMerged4.plot(axes=ax4, ignorePixelMask=256, show=False) ax4.set_ylim(-1e5, 1e6) ax4.set_title(f'{visit} (SM{spectrograph})') plt.savefig(f'trace_pfsMerged.pdf') plt.savefig(f'trace_pfsMerged.png')