-
Type:
Task
-
Status: Done (View Workflow)
-
Priority:
Normal
-
Resolution: Done
-
Affects Version/s: None
-
Fix Version/s: None
-
Component/s: None
-
Labels:None
-
Story Points:5
-
Sprint:2DDRP-2019 D, 2DDRP-2019 E, 2DDRP-2019 F
The purpose of this ticket is to document the post-stamp cutouts that I am using to study wavefront aberrations and create 2d PSF (e.g., PIPE2D-351 and the ticket for investigation of the February data that I still have to create). I believe this might be also useful for Keigo Nakamura and PIPE2D-355.
I have placed the data at /tigress/HSC/PFS/datacuts_Neven
When uncompressed, there are two folders 1. Dataframes, 2. Stamps_Cleaned.
************************
Dataframes can be opened as, after changing the path from
with your own path/Users/nevencaplar/Documents/PFS/ReducedData/
with open('/Users/nevencaplar/Documents/PFS/ReducedData/Data_Feb_5/Dataframes/finalNe_Feb2019.pkl', 'rb') as f:
finalNe_Feb2019=pickle.load(f)
or
with open('/Users/nevencaplar/Documents/PFS/ReducedData/Data_Feb_5/Dataframes/finalHgAr_Feb2019.pkl', 'rb') as f:
finalHgAr_Feb2019=pickle.load(f)
This dataframe contains information:
1. fiber number, as counted from the left,
2. x position of the spot on the detector,
3. y position on the detector,
4. wavelength of the spot,
5. is there another line close by that might compromise this line. Value 0=yes (i.e., use this line with caution), value 1= no (more confidence in this line),
6. lamp which was used,
7. ``effective'' x value on chip, after taking into account that there is roughly 69 pixels between two ccds.
************************
Stamps_Cleaned is where cutouts live. All of the images are stacked. The naming convention is
type+observation_number+index_of_the_spot+lamp+description+.npy
1. type: sci for cut from science image, mask for cut from the mask image, var for the cut from the variance image
2. observation number. For HgAr the values are [11796,11790,11784,11778,11772,11766,11760,11754,11748,11694,11700,11706,11712,11718,11724,11730,11736] and correspond to ['m4','m35','m3','m25','m2','m15','m1','m05','0','p05','p1','p15','p2','p25','p3','p35','p4'], i.e., moving from minus 4 (m4) to plus 4 (p4) defocus. For Neon the corresponding values are [12403,12397,12391,12385,12379,12373,12367,12361,12355,12355,12349,12343,12337,12331,12325,12319,12313,12307]
3.index_of_the_spot as deduced from the dataframes explained above
4. lamp - HgAr or Neon (Krypton in the future also)
5. description - Stacked or Stacked_Large. Stacked_Large gives a bit larger view of the spot.
So for example, one of the names is mask117666HgAr_Stacked.npy, describing mask of the data taken with HgAr, observation 11766 which correspond to data when the slit was at 'm15' position, i.e., -1.5 mm, and the spot is number 66 in the finalHgAr_Feb2019.pkl dataframe.
Some challenges remain, especially regarding cleaning the continuum from the HgAr data. I hope that once when we get reduceArc working (PIPE2D-375), this can be sorted out.
I am attaching two files
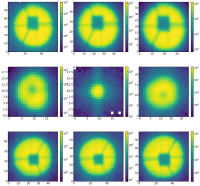
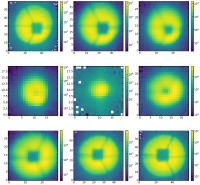
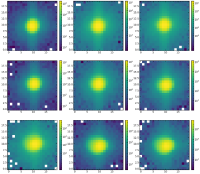
1. AsFunctionOfDifferentFibers.png showing the cutouts in the focus at the same wavelength but in different fibers
2. AsFunctionOfDefocus.png showing the cutouts of the same spot as a function of the defocus
-------------------------------------------------------------------------------------------------------------------------------------
Update, June 11:
I have added Data_May_28 in the folder (/tigress/HSC/PFS/datacuts_Neven). This contains the data taken with F/2.8 stops. The structure is the same as described above. HgAr data spans visits (17017 - 17130) and Neon data spans visits (16238 - 16351). Krypton and Xenon data still not included. In near future I hope to add F/2.5 data as well. These new datacuts include the cleaning from PIPE2D-401.
Below, I have also added an image showing an example of the cutouts as a function of defocus.
-------------------------------------------------------------------------------------------------------------------------------------
Update, June 25:
I have added Data_Jun 25 in the folder (/tigress/HSC/PFS/datacuts_Neven). This contains the data taken with F/2.5 stops. The structure is the same as described above. HgAr data spans visits (19238 - 19351) and Neon data spans visits (19472 - 19585). Krypton and Xenon data still not included. These new datacuts include the cleaning from PIPE2D-401.
- is blocked by
-
PIPE2D-401 Removal of HgAr continuum in focus and defocus
-
- Done
-
- relates to
-
PIPE2D-399 Create the postage stamps for the new LAM data (April 2019)
-
- Done
-
-
PIPE2D-351 Model the 2D PSF
-
- Done
-
-
PIPE2D-375 Investigate reduceArc failures on recent LAM data
-
- Done
-
-
PIPE2D-355 Model the 2D PSF using PSFEx
-
- Won't Fix
-